Online Experiments with a Bayesian Lens
From conversion metrics to posterior intuition
RCTBayesianA/B TestingPython
RCTBayesianA/B TestingPython
Online experiments (A/B tests and randomized controlled trials) help us compare a control and a treatment fairly by randomizing who sees what. A Bayesian lens keeps uncertainty front‑and‑center: as data arrives, we update beliefs and read posteriors as degrees of confidence about effects.
Start with the decision you want to make. Prefer simple, attributable metrics (e.g., conversion) and reach for composites only when they reflect a clear trade‑off (e.g., a weighted blend of sign‑ups and cancellations). Write down guardrails up front (e.g., latency, error rate) so wins don’t come at the wrong cost.
Beta prior.Conversion is the share of users who take an action (e.g., free → paid). With a Beta prior and Bernoulli likelihood, observing successes and failures simply updates the Beta’s parameters—the posterior stays in the same family and is easy to interpret.
Beta(a, b) lives on [0,1]. Its mean is a/(a+b). Larger (a+b) means more information and a tighter distribution. With a uniform prior Beta(1,1), seeing s successes and f failures yields Beta(1+s, 1+f).
![Beta(a=2, b=8) density on [0,1]](/articles/online-experiments-bayesian/assets/beta_ab.png)
import math
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
def beta_pdf(x, a, b):
x = min(max(x, 1e-6), 1-1e-6)
lg = math.lgamma
logB = lg(a) + lg(b) - lg(a+b)
logpdf = (a-1)*math.log(x) + (b-1)*math.log(1-x) - logB
return math.exp(logpdf)
xs = [i/1000 for i in range(0,1001)]
ys = [beta_pdf(x, 2, 8) for x in xs]
with plt.xkcd():
fig, ax = plt.subplots(figsize=(8,4))
ax.fill_between(xs, ys, color=(30/255,100/255,220/255), alpha=0.4)
ax.plot(xs, ys, color=(30/255,100/255,220/255), linewidth=2)
ax.set_xlim(0,1)
ax.set_ylim(0, max(ys)*1.1)
ax.set_title('Beta(a=2, b=8)')
ax.set_xlabel('p')
ax.set_ylabel('density')
fig.tight_layout()
fig.savefig('beta_ab.png', dpi=120)
print('wrote beta_ab.png')
When comparing variants, we often summarize control and treatment as Beta(a_c, b_c) and Beta(a_t, b_t). Overlap shows uncertainty; less overlap means clearer separation. The mean of each is a/(a+b).
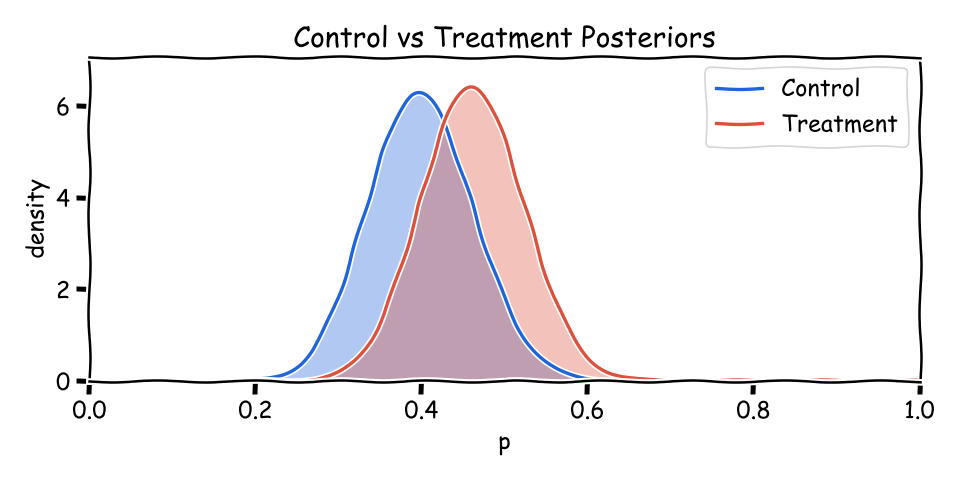
import math
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
def beta_pdf(x, a, b):
x = min(max(x, 1e-6), 1-1e-6)
lg = math.lgamma
logB = lg(a) + lg(b) - lg(a+b)
logpdf = (a-1)*math.log(x) + (b-1)*math.log(1-x) - logB
return math.exp(logpdf)
xs = [i/1000 for i in range(0,1001)]
y1 = [beta_pdf(x, 24, 36) for x in xs]
y2 = [beta_pdf(x, 30, 35) for x in xs]
with plt.xkcd():
fig, ax = plt.subplots(figsize=(8,4))
ax.fill_between(xs, y1, color=(30/255,100/255,220/255), alpha=0.35)
ax.fill_between(xs, y2, color=(220/255,80/255,60/255), alpha=0.35)
ax.plot(xs, y1, color=(30/255,100/255,220/255), linewidth=2, label='Control')
ax.plot(xs, y2, color=(220/255,80/255,60/255), linewidth=2, label='Treatment')
ax.set_xlim(0,1)
ax.set_ylim(0, max(max(y1), max(y2))*1.1)
ax.set_title('Control vs Treatment Posteriors')
ax.set_xlabel('p')
ax.set_ylabel('density')
ax.legend()
fig.tight_layout()
fig.savefig('two_posteriors.png', dpi=120)
print('wrote two_posteriors.png')
As more users arrive, (a+b) grows and each posterior narrows. The means can stay similar while uncertainty shrinks—that’s exactly what you want from accumulating evidence.
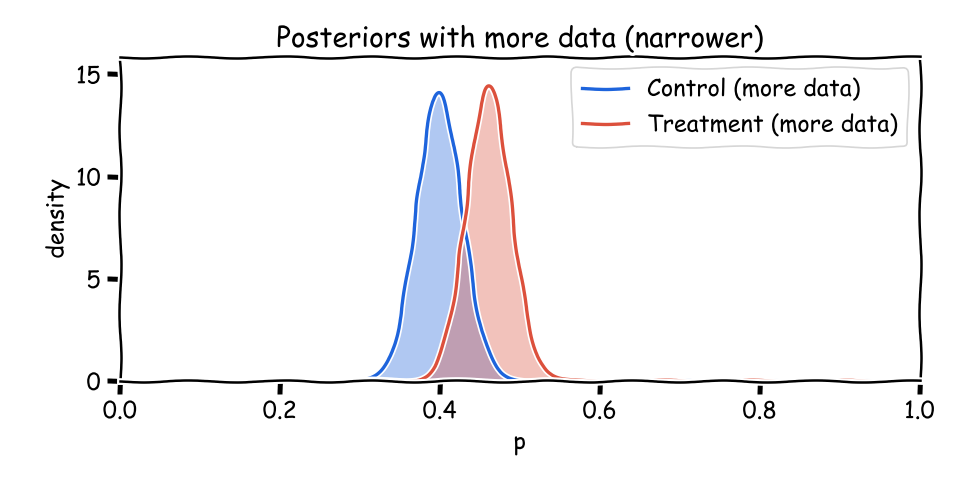
import math
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
def beta_pdf(x, a, b):
x = min(max(x, 1e-6), 1-1e-6)
lg = math.lgamma
logB = lg(a) + lg(b) - lg(a+b)
logpdf = (a-1)*math.log(x) + (b-1)*math.log(1-x) - logB
return math.exp(logpdf)
xs = [i/1000 for i in range(0,1001)]
y1 = [beta_pdf(x, 24*5, 36*5) for x in xs]
y2 = [beta_pdf(x, 30*5, 35*5) for x in xs]
with plt.xkcd():
fig, ax = plt.subplots(figsize=(8,4))
ax.fill_between(xs, y1, color=(30/255,100/255,220/255), alpha=0.35)
ax.fill_between(xs, y2, color=(220/255,80/255,60/255), alpha=0.35)
ax.plot(xs, y1, color=(30/255,100/255,220/255), linewidth=2, label='Control (more data)')
ax.plot(xs, y2, color=(220/255,80/255,60/255), linewidth=2, label='Treatment (more data)')
ax.set_xlim(0,1)
ax.set_ylim(0, max(max(y1), max(y2))*1.1)
ax.set_title('Posteriors with more data (narrower)')
ax.set_xlabel('p')
ax.set_ylabel('density')
ax.legend()
fig.tight_layout()
fig.savefig('narrowing.png', dpi=120)
print('wrote narrowing.png')
When the curves overlap, both variants remain plausible. Report the probability that treatment beats control, plus a credible interval for the difference. These are honest statements about uncertainty—not all‑or‑nothing verdicts.
A friendly way to summarize is by simulation: draw many samples from each posterior, compute the lift (p_treat − p_control), then report P(lift > 0) and a credible interval. This Monte Carlo view turns curves into numbers you can act on.
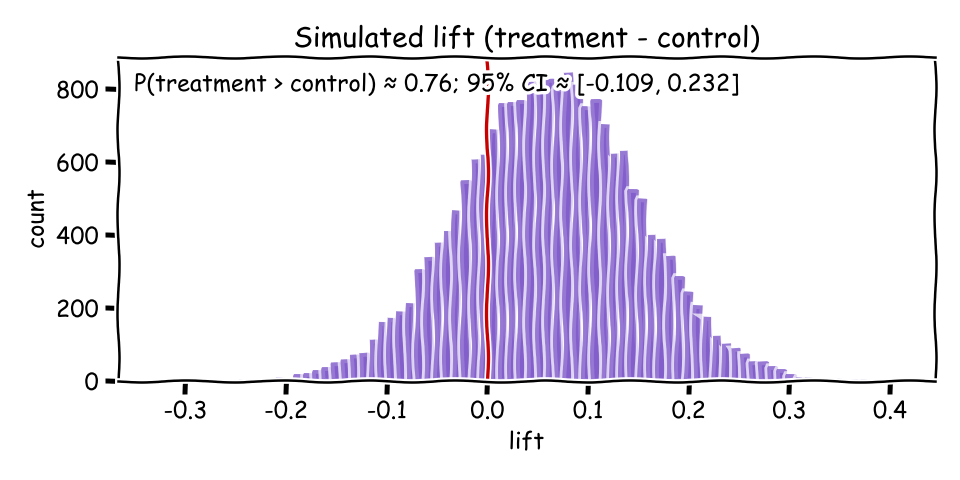
import random, math
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
# Posterior parameters (match earlier example)
a1,b1 = 24,36 # control
a2,b2 = 30,35 # treatment
N = 20000
samples = []
for _ in range(N):
pc = random.betavariate(a1, b1)
pt = random.betavariate(a2, b2)
samples.append(pt - pc)
p_better = sum(1 for x in samples if x > 0) / N
lo, hi = sorted(samples)[int(0.025*N)], sorted(samples)[int(0.975*N)]
with plt.xkcd():
fig, ax = plt.subplots(figsize=(8,4))
ax.hist(samples, bins=80, color=(120/255,80/255,200/255), alpha=0.7)
ax.axvline(0.0, color=(200/255,0,0), linewidth=2)
ax.set_title('Simulated lift (treatment - control)')
ax.set_xlabel('lift')
ax.set_ylabel('count')
msg = f"P(treatment > control) ≈ {p_better:.2f}; 95% CI ≈ [{lo:.3f}, {hi:.3f}]"
ax.text(0.02, 0.95, msg, transform=ax.transAxes, va='top')
fig.tight_layout()
fig.savefig('delta_hist.png', dpi=120)
print('wrote delta_hist.png')
print(msg)
P(treatment > control) and a credible interval for the lift.A Bayesian approach keeps uncertainty visible from start to finish. By modeling conversion with Beta posteriors, visualizing overlap, and simulating lift, you can make measured, decision‑ready calls grounded in data—not just a p‑value.
If you're exploring related work and need hands‑on help, I'm open to consulting and advisory. Contact me on LinkedIn.